本文由医信融合团队成员“陈浩然”撰写,已同步至微信公众号“医信融合创新沙龙”与“研究生学生信”,更多精彩内容欢迎关注!


名人名言
沃兹基曾经说过:复杂的劳动是为了简单的重复

Article name:A comprehensive transcriptome signature of murine hematopoietic stem cell aging
Journal:blood
Doi:10.1182/blood.2020009729
IF:23.629
Position:Figure 1C
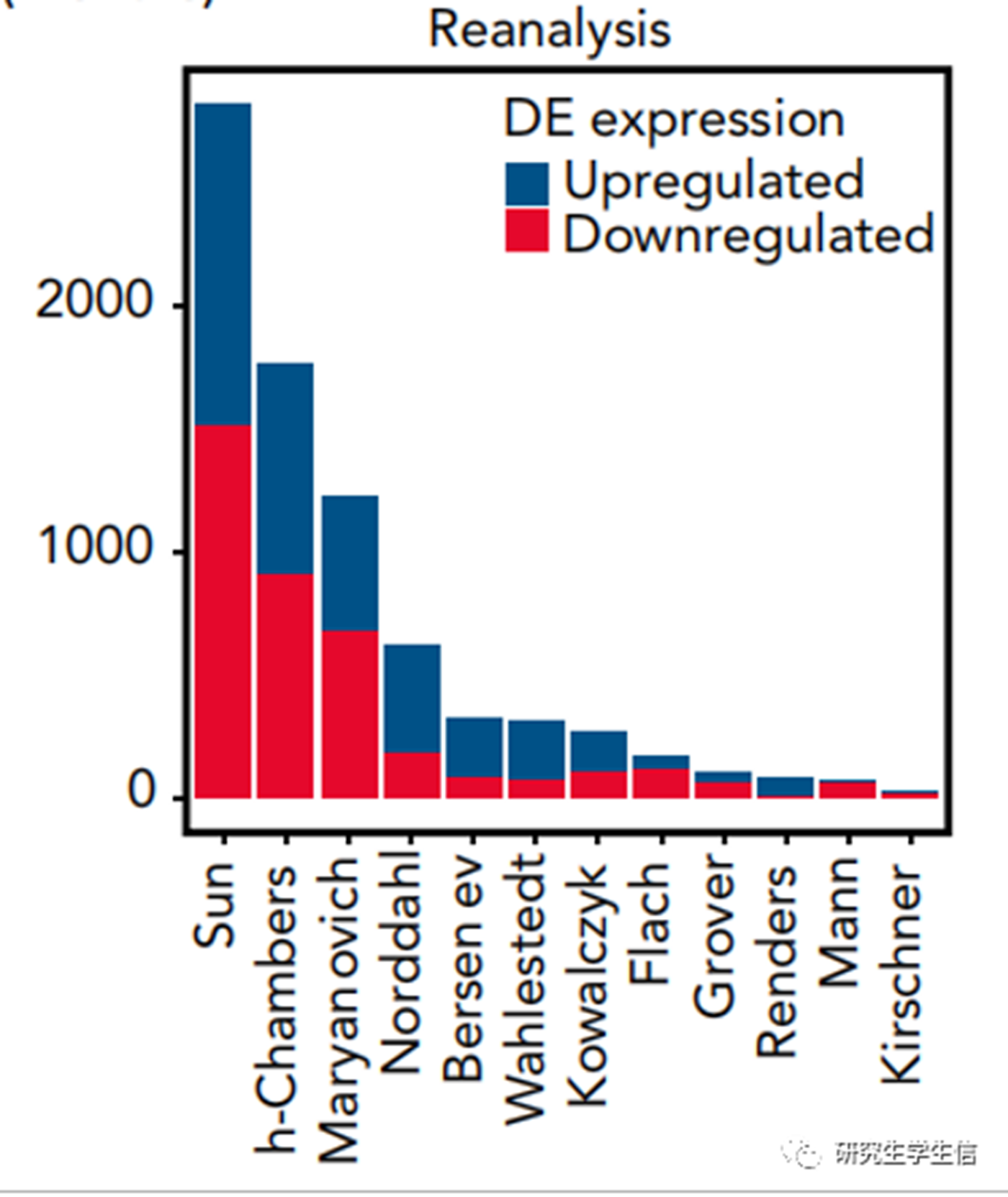
这是一张简单的条形图,用鼠标比着尺子画的话,10分钟就能画完,但是如果用R的话,用了俩小时

不过学习了一些东西,也算傻人有傻福:
• 12个数据集的系统性规范化整理
• 对于建库过程中处理数据时的标准化问题
• R语言中list的批量合并
• 管道运算
• 重复基因的分组与去重
• 去除空值
• 探针与基因中一对多的处理方法
• ggplot2中如何按照y轴值的大小顺序绘制
• 绘制重叠条形图
• 如何选取好看的颜色
• y轴坐标轴标签如何修改
• 如何去除背景刻度和背景颜色
• y轴标签如何旋转一定角度并且紧贴坐标轴
• 标题,坐标轴标签如何修改
• 整个图片主题字体如何修改
由于太晚了,就以两种方式进行分享:
1:可以在完整阅读文献后,下载原始数据,参考以下代码进行运行
library(ggplot2)
library(dplyr)
library(tidyr)
library(reshape)
rm(list = ls())
setwd("./file")
Bersenev <- read.table("Bersenev_GSE39553.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%
.[,c("Gene.symbol","logFC")]
colnames(Bersenev)<-c("genes","Bersenev")
Chambers <- read.table("Chambers_GSE6503.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene.symbol","logFC")]
colnames(Chambers)<-c("genes","Chambers")
Flach <- read.table("Flach_GSE48893.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene.symbol","logFC")]
colnames(Flach)<-c("genes","Flach")
Grover <- read.table("Grover_GSE70657.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene","avg_logFC")]
colnames(Grover)<-c("genes","Grover")
Kirschner <- read.table("Kirschner_GSE87631.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene","avg_logFC")]
colnames(Kirschner)<-c("genes","Kirschner")
Kowalczyk <- read.table("Kowalczyk_GSE59114.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene","avg_logFC")]
colnames(Kowalczyk)<-c("genes","Kowalczyk")
Lazare <- read.table("Lazare_GSE128050.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("external_gene_name","logFC")]
colnames(Lazare)<-c("genes","Lazare")
Mann <- read.table("Mann_GSE1004426.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene","avg_logFC")]
colnames(Mann)<-c("genes","Mann")
Maryanovich <- read.table("Maryanovich_GSE109546.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("external_gene_name","logFC")]
colnames(Maryanovich)<-c("genes","Maryanovich")
Norddahl <- read.table("Norddahl_GSE27686.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene.symbol","logFC")]
colnames(Norddahl)<-c("genes","Norddahl")
Sun <- read.table("Sun_GSE47817.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("external_gene_name","logFC")]
colnames(Sun)<-c("genes","Sun")
Wahlestedt <- read.table("Wahlestedt_GSE44923.csv",sep = "\t",
header = T,skip = 1,quote = "")%>%.[,c("Gene.symbol","logFC")]
colnames(Wahlestedt)<-c("genes","Wahlestedt")
all_data <- list(Bersenev,Chambers,Flach,Grover,Kirschner,
Kowalczyk,Lazare, Mann,Maryanovich,Norddahl,
Sun,Wahlestedt)
all_pub <- purrr::reduce(.x = all_data,.f = full_join,by="genes")
geneMatrix <- all_pub %>% group_by(genes) %>% filter (!duplicated(genes))
geneMatrix<-geneMatrix[geneMatrix[,1]!="",]
blood_output<-separate_rows(geneMatrix,genes,sep = "///")
write.table(blood_output,"output.txt",sep = "\t",quote = F,row.names = F,col.names = T)
data <- blood_output[,2:13]
plot_matrix <- matrix(nrow = ncol(data),ncol = 3,dimnames = list(NULL,c("name","Upregulated","Downregulated")))
for (x in 1:ncol(data)) {
data_name <- colnames(data)[x]
non_na_data <- na.omit(data[,x])
Upregulated <- length(non_na_data[non_na_data>0])
Downregulated <- length(non_na_data[non_na_data<0])
plot_matrix[x,] <- c(data_name,Upregulated,Downregulated)
}
plot_matrix <- as.data.frame(plot_matrix)
plot_matrix <- melt(plot_matrix,id.vars = c("name"))
plot_matrix$value <- as.numeric(plot_matrix$value)
pl <- ggplot(data=plot_matrix, aes(x=reorder(name,-value), y=value)) +
geom_bar(stat = "identity",aes(fill=variable))+
scale_fill_manual(values=c("#005187","#e5082c"))+
scale_y_continuous(breaks=c(1000,2000,3000),
labels=c("1000", "2000", "3000"))+
theme_bw()+
theme(panel.grid=element_blank())+
theme(axis.text.x = element_text(angle = 90, hjust = 1, vjust = 1))+
labs(x="",y="# of reported DE genes",title = "Reanalysis")+
theme(text = element_text(family = "Arial",face = "bold"))
ggsave(pl, filename = "blood_figure_1c.pdf", device = cairo_pdf,
width = 8, height = 7, units = "in")
2:后台回复blood1c领取代码和数据,整个代码和文件将以project形式发送,也就是说,将文件解压后:
1. 双击blood_figure1.Rproj
1. 打开code文件夹中的code.R
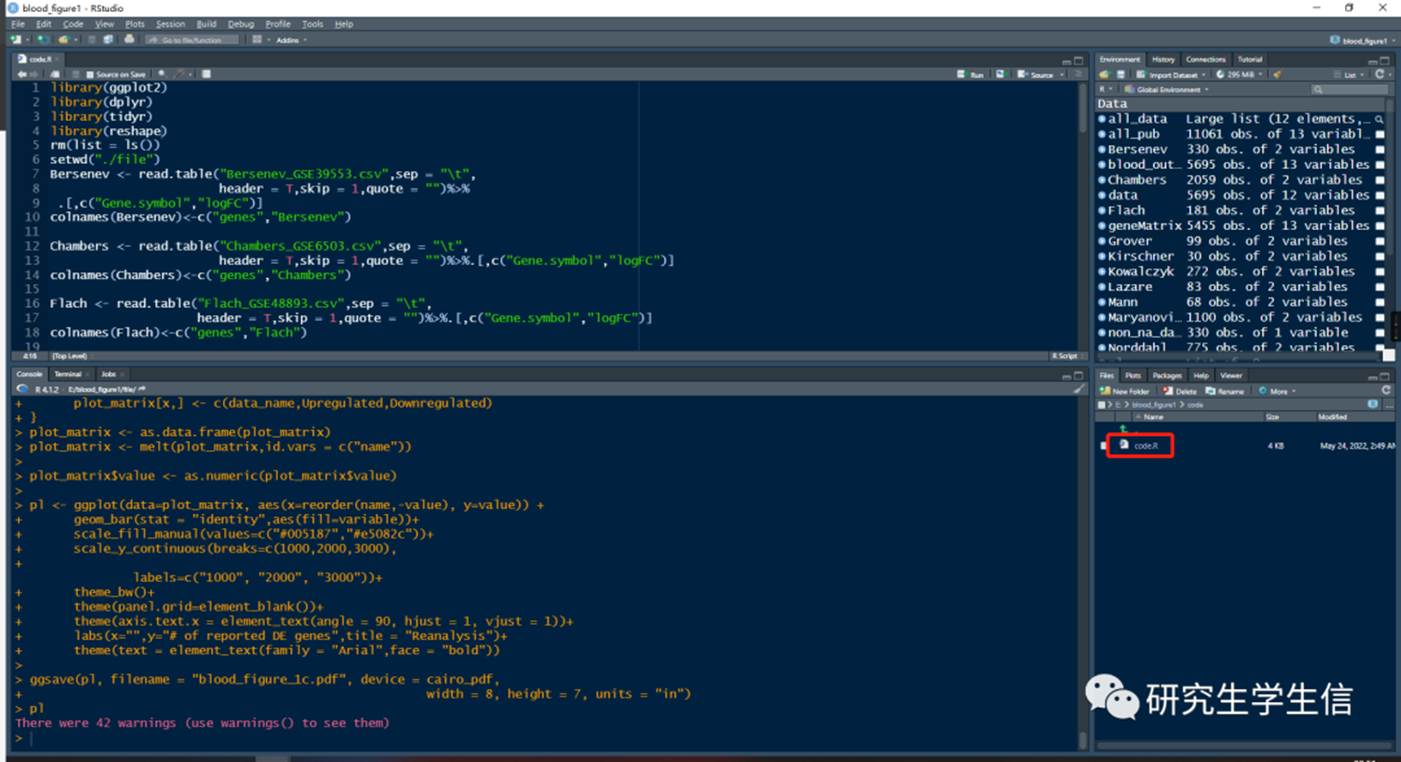
1. 全选、运行即可
1. 结果将保存在file文件夹中,也会在Plots窗口展示
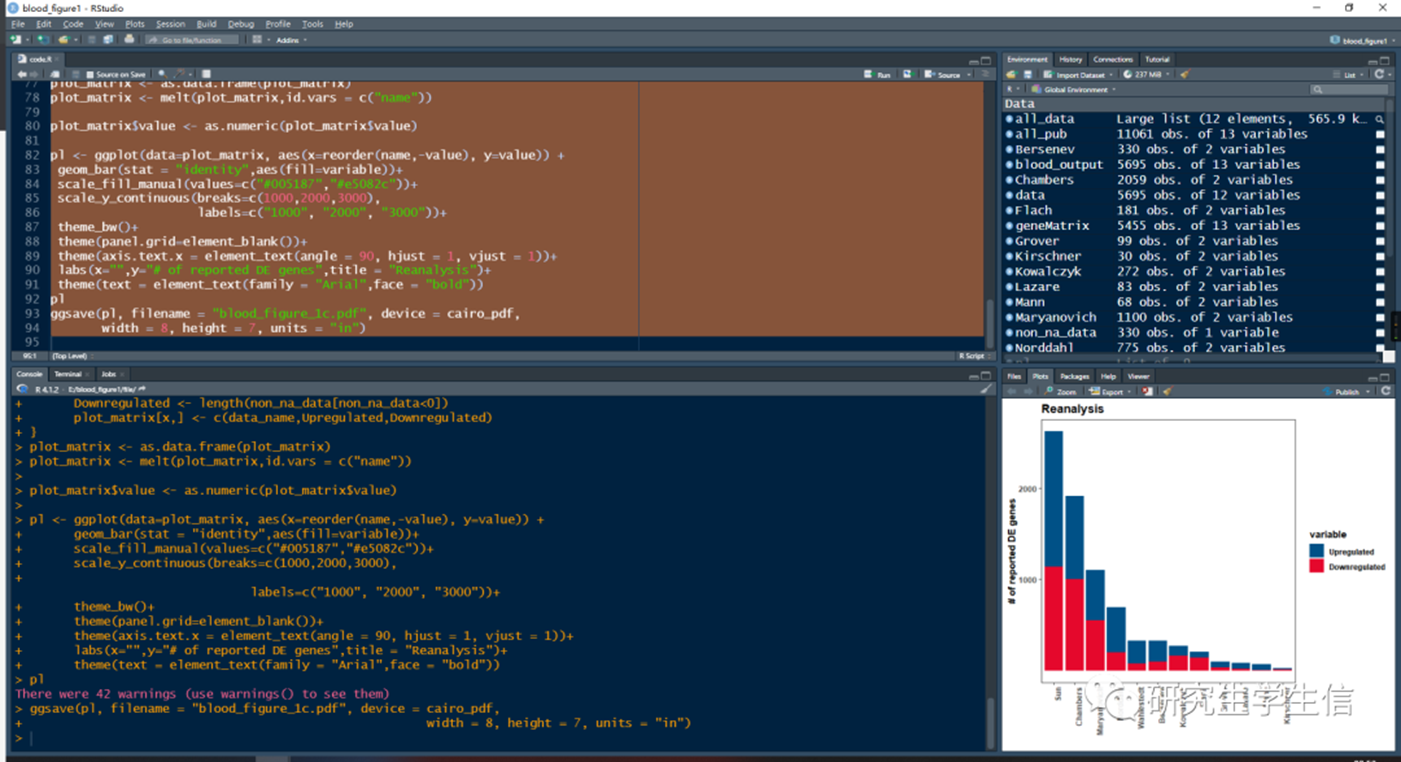
那么,如何才能更加美化一下图片呢?说不定以后就更新了
对于代码,可以私信交流
图文:陈浩然
本文编辑:莫状