本文由医信融合团队成员“张皓旻”撰写,已同步至微信公众号“医信融合创新沙龙”,更多精彩内容欢迎关注!

火山图是最常见的生物信息可视化图片,今天我们一起来画一张高颜值火山图!!!(文末附完整代码,快点收藏吧)
前期准备
输入数据格式
输入数据是差异分析获得的所有结果文件,我们看一下具体的内容。

安装需要的软件包
#在R语言中运行
install.packages("ggplot2")
install.packages("ggrepel")
开始绘图
导入软件包、读入数据、设置工作目录、设置差异基因阈值
library(ggplot2)
library(ggrepel)
setwd("J:\\code\\火山图")
dataset <- read.table('volcano.txt',header = TRUE)
cut_off_pvalue = 0.05
cut_off_logFC = 0.8
绘图
##定义差异基因
dataset$change = ifelse(dataset$P.Value < cut_off_pvalue & abs(dataset$logFC) >= cut_off_logFC,
ifelse(dataset$logFC> cut_off_logFC ,'Up','Down'),
'Stable')
##绘图
pdf("volcanol_pvalue.pdf")
ggplot(
dataset,
aes(x = logFC,
y = -log10(P.Value),
colour=change)) +
geom_point(alpha=0.4, size=3.5) +
scale_color_manual(values=c("#546de5", "#d2dae2","#ff4757"))+
geom_vline(xintercept=c(-1,1),lty=4,col="black",lwd=0.8) +
geom_hline(yintercept = -log10(cut_off_pvalue),lty=4,col="black",lwd=0.8) +
labs(x="log2(fold change)",y="-log10 (p-value)")+
theme_bw()+
theme(plot.title = element_text(hjust = 0.5),
legend.position="right",
legend.title = element_blank()
)
dev.off()
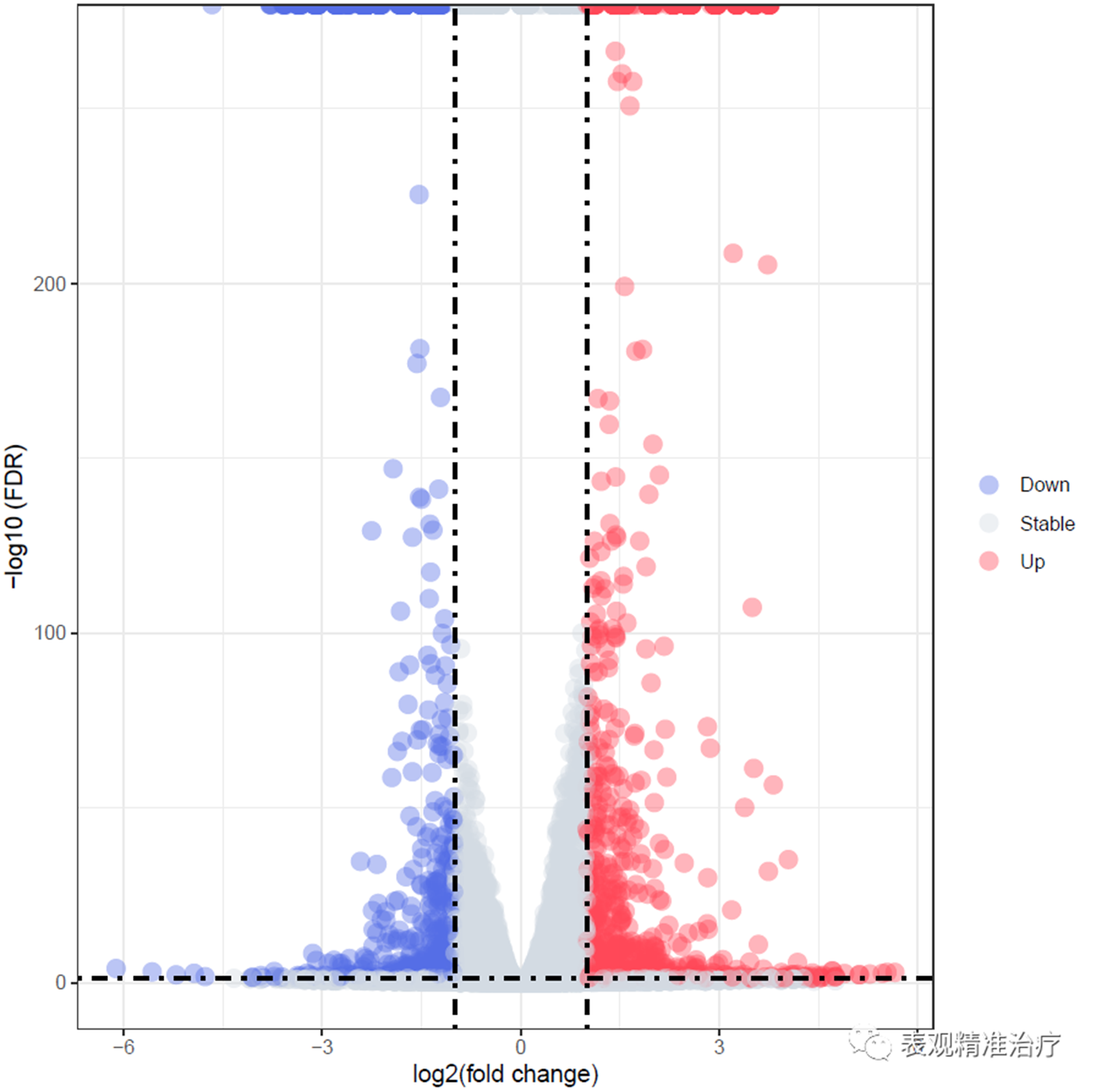
如果需要标注基因
dataset$label = ifelse(dataset$P.Value < cut_off_pvalue & abs(dataset$logFC) >= 3, as.character(dataset$gene),"")
p+geom_text_repel(data = dataset, aes(x = dataset$logFC,
y = -log10(dataset$P.Value),
label = label),
size = 3,box.padding = unit(0.8, "lines"),
point.padding = unit(0.8, "lines"),
show.legend = FALSE)
完整代码
绘制火山图(FDR筛选)
library(ggplot2)
cut_off_stat = 0.05 #设置统计值阈值
cut_off_logFC = 1 #设置表达量阈值,可修改
diff_res[is.na(diff_res)] <- 0
diff_res$change = ifelse(diff_res$padj < cut_off_stat & abs(diff_res$log2FoldChange) >= cut_off_logFC,
ifelse(diff_res$log2FoldChange> cut_off_logFC ,'Up','Down'),
'Stable')
pdf("volcanol_FDR.pdf")
ggplot(diff_res,
aes(x = log2FoldChange,
y = -log10(padj),
colour=change)) +
geom_point(alpha=0.4, size=3.5) +
scale_color_manual(values=c("#546de5", "#d2dae2","#ff4757"))+
geom_vline(xintercept=c(-1,1),lty=4,col="black",lwd=0.8) +
geom_hline(yintercept = -log10(cut_off_stat),lty=4,col="black",lwd=0.8) +
labs(x="log2(fold change)",
y="-log10 (FDR)")+
theme_bw()+
theme(plot.title = element_text(hjust = 0.5),
legend.position="right",
legend.title = element_blank()
)
dev.off()
绘制火山图(pvalue筛选)
library(ggplot2)
cut_off_stat = 0.05 #设置统计值阈值
cut_off_logFC = 1 #设置表达量阈值,可修改
diff_res[is.na(diff_res)] <- 0
diff_res$change = ifelse(diff_res$pvalue < cut_off_stat & abs(diff_res$log2FoldChange) >= cut_off_logFC,
ifelse(diff_res$log2FoldChange> cut_off_logFC ,'Up','Down'),
'Stable')
pdf("volcanol_pvalue.pdf")
ggplot(diff_res,
aes(x = log2FoldChange,
y = -log10(pvalue),
colour=change)) +
geom_point(alpha=0.4, size=3.5) +
scale_color_manual(values=c("#546de5", "#d2dae2","#ff4757"))+
geom_vline(xintercept=c(-1,1),lty=4,col="black",lwd=0.8) +
geom_hline(yintercept = -log10(cut_off_stat),lty=4,col="black",lwd=0.8) +
labs(x="log2(fold change)",
y="-log10 (p-value)")+
theme_bw()+
theme(plot.title = element_text(hjust = 0.5),
legend.position="right",
legend.title = element_blank()
)
dev.off()
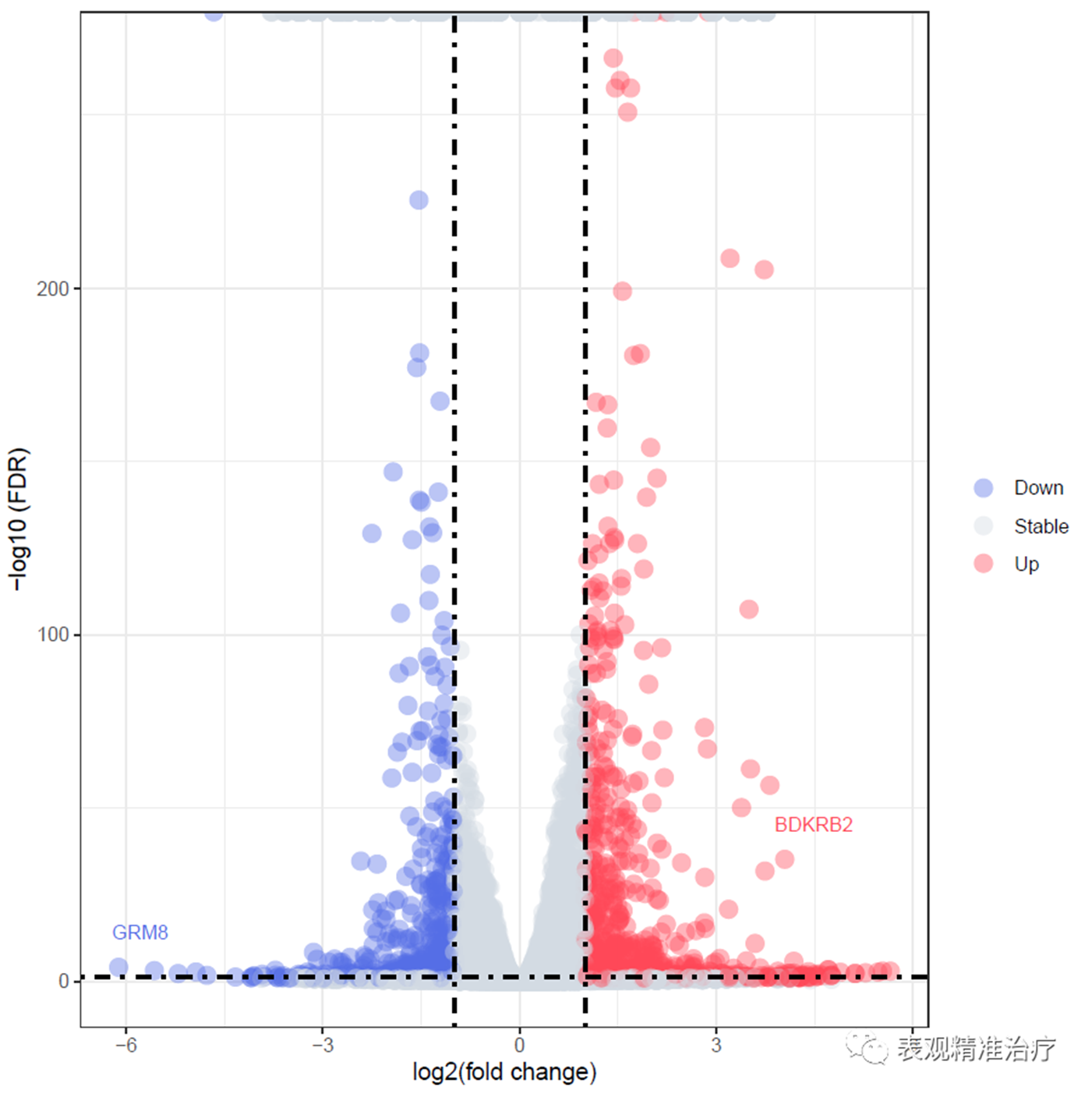
绘制带有基因名称的火山图
library(ggplot2)
library(ggrepel)
cut_off_stat = 0.05 #设置统计值阈值
cut_off_logFC = 1 #设置表达量阈值,可修改
diff_res[is.na(diff_res)] <- 0
diff_res$change = ifelse(diff_res$pvalue < cut_off_stat & abs(diff_res$log2FoldChange) >= cut_off_logFC,
ifelse(diff_res$log2FoldChange> cut_off_logFC ,'Up','Down'),
'Stable')
diff_res$label = ifelse(diff_res$padj < cut_off_stat & abs(diff_res$log2FoldChange) >= 4, as.character(diff_res$gene_id),"")
pdf("volcanol_gene.pdf")
ggplot(diff_res,
aes(x = log2FoldChange,
y = -log10(padj),
colour=change)) +
geom_point(alpha=0.4, size=3.5) +
scale_color_manual(values=c("#546de5", "#d2dae2","#ff4757"))+
geom_vline(xintercept=c(-1,1),lty=4,col="black",lwd=0.8) +
geom_hline(yintercept = -log10(cut_off_stat),lty=4,col="black",lwd=0.8) +
labs(x="log2(fold change)",
y="-log10 (FDR)")+
theme_bw()+
theme(plot.title = element_text(hjust = 0.5),
legend.position="right",
legend.title = element_blank()
)+
geom_text_repel(data = diff_res, aes(x = diff_res$log2FoldChange,
y = -log10(diff_res$padj),
label = label),
size = 3,box.padding = unit(0.8, "lines"),
point.padding = unit(0.8, "lines"),
show.legend = FALSE)
dev.off()
图文:张皓旻
本文编辑:李新龙